The biological phenomena are governed by complex network systems including many species of molecules, cells or organs. For the aim of understanding the functions of complex systems, we adopt mathematical and computational methods.
By theoretical approaches we decipher huge amounts of experimental information, and give integrative understanding for the biological systems.
Our final goal is to open a new bioscience which will progress by the repeats of the theoretical predictions and the experimental verifications.
RESEARCH
-
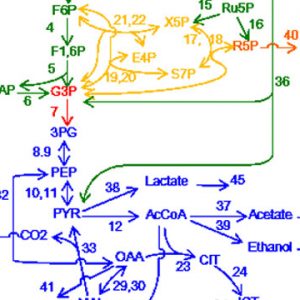
Complete control of a gene regulatory network of ascidian embryo by a few factors identified by a mathematical theory.
VIEW MORE -
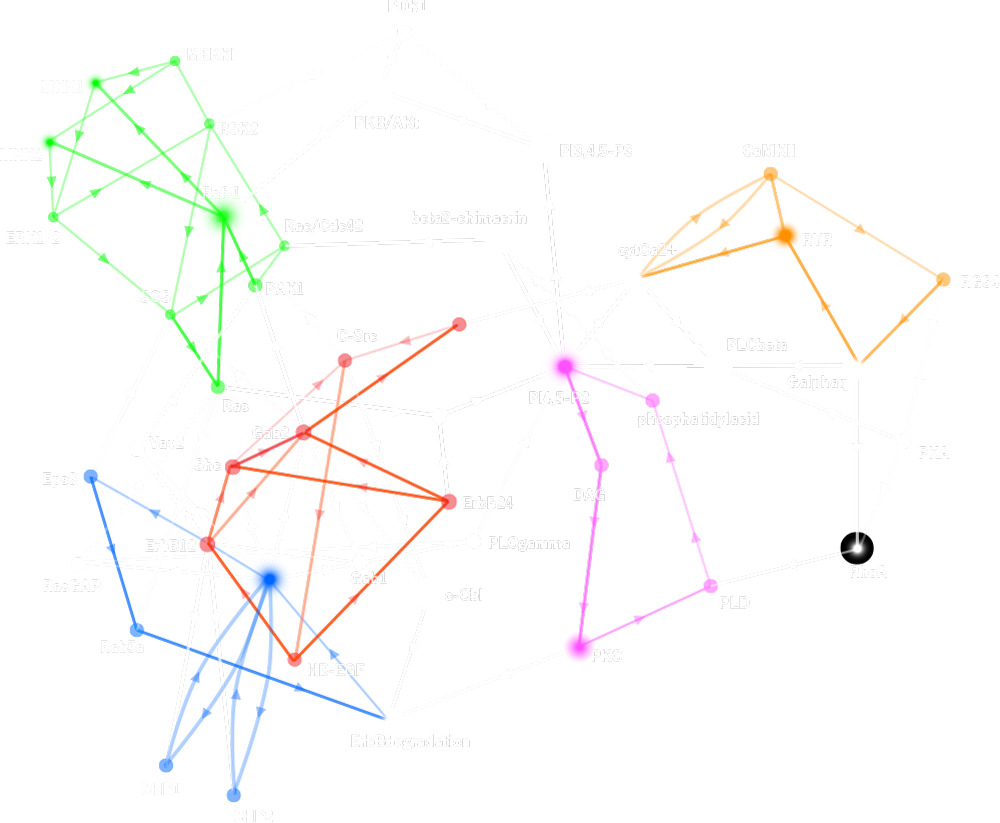
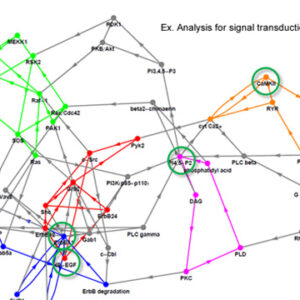
Determining responses of chemical reaction systems from structure of networks
VIEW MORE -
Structure and dynamics of regulatory networks
VIEW MORE -

Physics of organelle morphogenesis
VIEW MORE
NEWS
- publication
Okada, T., Isacchini, G., Yu, Q., & Hallatschek, O. (2025). Uncovering heterogeneous intercommunity disease transmission from neutral allele frequency time series. Proceedings of the National Academy of Sciences, 122(48), e2500663122.
- publication
Huang, Y. J., Okada, T., & Mochizuki, A. (2025). Uncovering bifurcation behaviors of biochemical reaction systems from network topology. Scientific Reports, 15(1), 27596. We have developed a method for analyzing bifurcation phenomena based solely on the network structure information of chemical reaction networks.
- publication
Wang Z., Inoue Y., Mochizuki A. & Tsukaya H., (2025) Biregionally differentiated growth generates sharp apex and concave joints in leaves. Plant Journal 123 (1), e70310. e70310. doi: 10.1111/tpj.70310 Through joint research with the Tsukaya Laboratory at the University of Tokyo, we have elucidated the mechanism by which “pointed leaf tips” are formed. This research was introduced in the following press release.
- publication
Ooka H., Wintzer M.E., Komatsu H., Suda T., Adachi K., Li A., Kong S., Hashizume D., Mochizuki A. and Nakamura R. (2024) Microkinetic model to rationalize the lifetime of electrocatalysis: trade-off between activity and stability. Journal of Physical Chemistry Letters 15(40), 10079–10085.
- publication
Okino R., Mukai K., Oguri S., Masuda M., Watanabe S., Yoneyama Y., Nagaosa S., Miyamoto T., Mochizuki A., Takahashi S.-I. and Hakuno F. (2024) IGF-I concentration determines cell fate by converting signaling dynamics as a bifurcation parameter in L6 myoblasts. Scientific Reports 14, 20699.
- publication
Okada, T., Isacchini, G., Yu, Q., & Hallatschek, O. (2025). Uncovering heterogeneous intercommunity disease transmission from neutral allele frequency time series. Proceedings of the National Academy of Sciences, 122(48), e2500663122.
- publication
Huang, Y. J., Okada, T., & Mochizuki, A. (2025). Uncovering bifurcation behaviors of biochemical reaction systems from network topology. Scientific Reports, 15(1), 27596. We have developed a method for analyzing bifurcation phenomena based solely on the network structure information of chemical reaction networks.
- publication
Wang Z., Inoue Y., Mochizuki A. & Tsukaya H., (2025) Biregionally differentiated growth generates sharp apex and concave joints in leaves. Plant Journal 123 (1), e70310. e70310. doi: 10.1111/tpj.70310 Through joint research with the Tsukaya Laboratory at the University of Tokyo, we have elucidated the mechanism by which “pointed leaf tips” are formed. This research was introduced in the following press release.
- publication
Ooka H., Wintzer M.E., Komatsu H., Suda T., Adachi K., Li A., Kong S., Hashizume D., Mochizuki A. and Nakamura R. (2024) Microkinetic model to rationalize the lifetime of electrocatalysis: trade-off between activity and stability. Journal of Physical Chemistry Letters 15(40), 10079–10085.
- publication
Okino R., Mukai K., Oguri S., Masuda M., Watanabe S., Yoneyama Y., Nagaosa S., Miyamoto T., Mochizuki A., Takahashi S.-I. and Hakuno F. (2024) IGF-I concentration determines cell fate by converting signaling dynamics as a bifurcation parameter in L6 myoblasts. Scientific Reports 14, 20699.