- HOME
- Research
- Present Works
- Physics of organelle morphogenesis
Present Works
Physics of organelle morphogenesis
Our cell (eukaryotic cell) has various organelles made by lipid bilayer membrane. They realize their specific functions. For example, Golgi apparatus modifies proteins and sorts them in order to transport them into adapted locations, mitochondrion produces energy carrying molecules from sugar and oxygen, and autophagosome emerges in starving conditions and depredates cellular components. The cooperation among organelles achieves complex behaviors of eukaryotic cells, which is not observed in prokaryotic cells.
The variety in shapes is another attractive feature of organelles. Since these shapes are considered to be tightly linked to their functions, studying how the shapes are made is fundamental subject in biology. However, there is a big difficulty in studying the organelle shapes; there is no way of observing the behaviors (morphological dynamics) of organelles, although their fixed shapes (snapshots of organelle shapes) are observable with electron microscopy. Therefore to understand the organelle behaviors, we have to extrapolate their dynamics from the observations on snapshots.
We approach this hard problem using physical modeling of membrane and computer simulations.
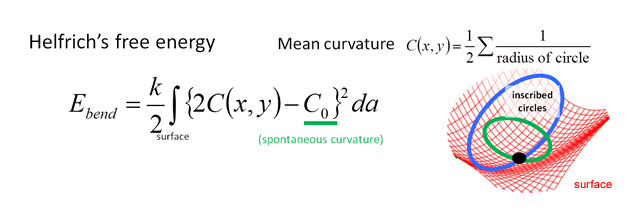
In physics, the motion of an object is calculated by giving the energy function of it. Since the membrane stores the bending energy, we estimate shape-associated energy by estimating the total bendedness of the membrane, and deformation motion is solved with this shape-stored energy. In addition, a complicated shaped membrane like organelle is realized with membrane-bending molecules (proteins and lipids). This effect is regarded as modification of bending energy at the presence of these molecules. Including these two fundamental effects, we made basic physical model of bio-membrane systems. Adding specific aspect of each organelle, we get physical model of each organelle.
However we have another difficulty in making specific organelle models; there are quite few information about membrane-bending molecules in organelles. Instead, the abundantly available data is snapshots of organelles. Therefore, in our study, we extrapolate the physical models of specific organelles based on the basic physical model and snapshots and try to understand total organelle behaviors using these specific models. No we are running three project
- physical modeling of reassembly process of Golgi apparatus,
- morphological response of caveolae to hypotonic shock
- molecule distributions which ensure the autophagosome formation.
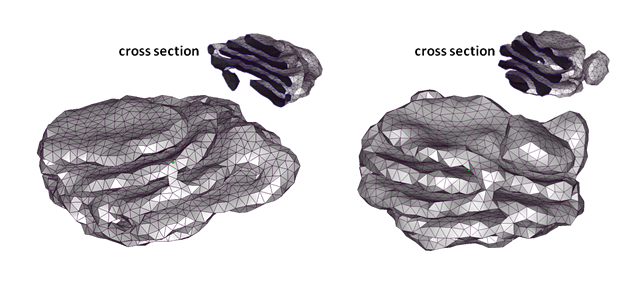
Computer simulation of Golgi reassembly model
Using polygon membrane model and Monte-Carlo method for membrane deformation dynamics, we reconstruct Golgi reassembly process and discuss the physical properties of it.