- HOME
- Research
- Present Works
- Understanding cell movements from image analysis
Present Works
Understanding cell movements from image analysis
During gastrulation in the mouse embryo, cells show dynamic movements.
These movments including epiblast invagination and mosodermal expansion lead to the establishment of the three-layered body plan.
However we do not have live images because of the phototoxicity and the complex morphology of the embryo.
To overcome these problems, Dr. Nonaka (NIBB, Japan) introduced new techniques with digital light sheet microscope (DSLM).
This new techniques can enable us to achieve deep and high time resolution live images.
We have got 4D live imaging data and achieved 3D tracking analysis and statistical analysis.
Revealed new findings are the followings:
- interkinetic nuclear migration (INM) occurs in the epiblast,
- primary driving force for INM at epiblast is not pressure from neighboring nuclei,
- mesodermal cells migrate not as a sheet but as individual cells without coordination.
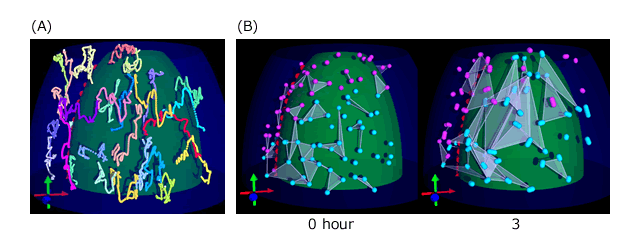
Three-dimensional tracking of mesodermal nuclei.
(A) Computationally reconstructed trajectories.White mark at the end of trajectory indicates the latest point.
(B) Comparison of triangles connected neighboring nuclei at first time point and after 3 hours. After 3 hours, the areas of the triangles become larger and some triangles overlap each other, indicating that nuclei in the mesoderm do not migrate in straight lines.